Tuning the preprocessing
In this notebook, we explain how to tune the preprocessing steps with available options in phyaat library. Basically two preprocessing steps are provided, filtering and artifact removal. For filtering, conventional way is to filter EEG with highpass filter (0.5 Hz), however you could choose any range. Second Artifact Removal, currently onle one method of artifact removal is implemented in phyaat, feel free to choose external libraries such as mne. The ICA based approach has a few hyperparameters, such as method to compute IC components, windowsize to process, threshold of kurtosis and correlation coefficient. We will see how to do that.
Table of Contents
Import libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import svm
import phyaat
print('Version :' ,phyaat.__version__)
import phyaat as ph
PhyAAt Processing lib Loaded...
Version : 0.0.2
#help(ph.Subject.getXy_eeg)
Read the data of subject=1
dirPath = ph.download_data(baseDir='../PhyAAt_Data', subject=1,verbose=0,overwrite=False)
baseDir='../PhyAAt_Data'
SubID = ph.ReadFilesPath(baseDir)
Subj = ph.Subject(SubID[1])
Total Subjects : 1
Filtering
With Custum frequency range
Subj.filter_EEG(band =[0.5,40],btype='bandpass',order=5)
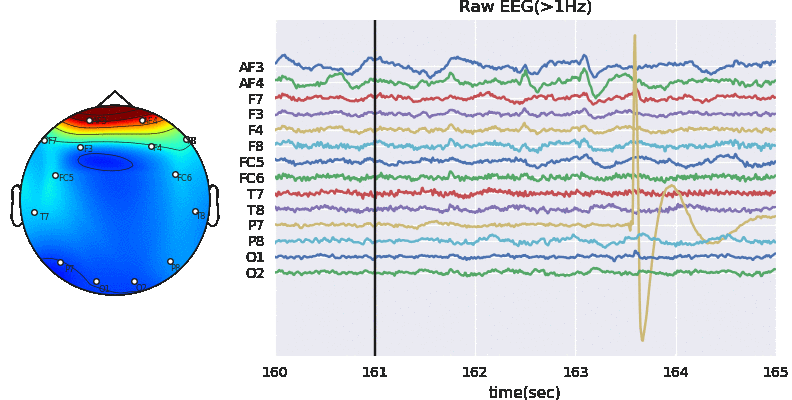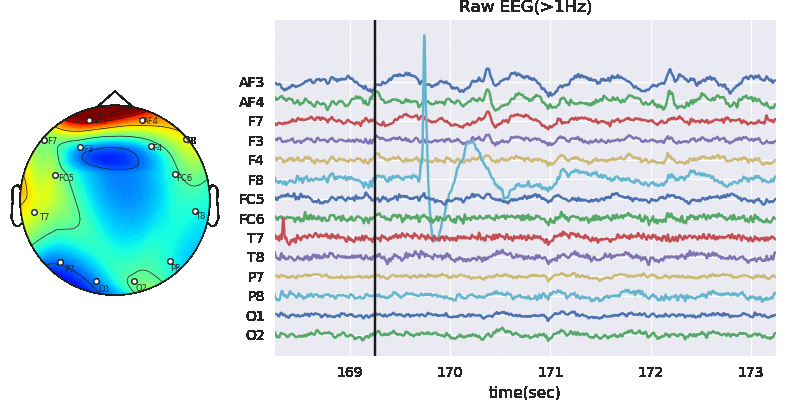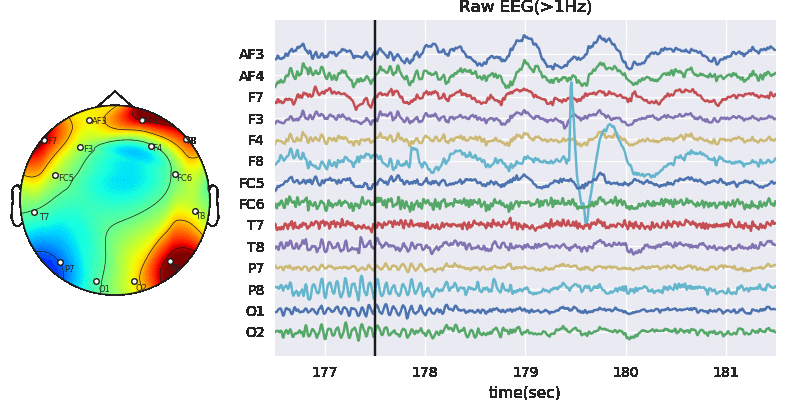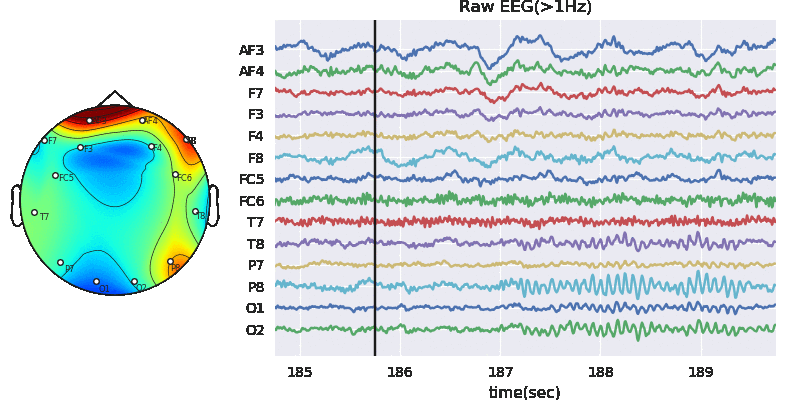
Artifact removal using ICA
Tune threshold, windowsize, ICA method
KurThr = 1.5
Corr = 0.7
ICAMed = 'infomax' #picard, fastICA
winsize=128*20 # 20sec
Subj.correct(method='ICA',winsize=winsize,hopesize=None,Corr=Corr,KurThr=KurThr,
ICAMed=ICAMed,verbose=1,
window=['hamming',True],winMeth='custom')
ICA Artifact Removal : infomax
100%|########################################################|
# Check help for details
help(ph.Subject.correct)
Help on function correct in module phyaat.ProcessingLib:
correct(self, method='ICA', winsize=128, hopesize=None, Corr=0.8, KurThr=2,
ICAMed='extended-infomax', verbose=0, window=['hamming', True], winMeth='custom')
method: 'ICA', ('WPA', 'ATAR' ) - not yet updated to library
ICAMed: ['fastICA','infomax','extended-infomax','picard']
winsize: 128, window size to processe
hopesize: 64, overlapping samples, if None, hopesize=winsize//2
window: ['hamming',True], window[1]=False to avoid windowing,
KurThr: (2) threshold on kurtosis to eliminate artifact,
ICA component with kurtosis above threshold are removed.
Corr = 0.8, correlation threshold, above which ica components are removed.
T3: Semanticity Classification
# setting task=-1, does extract the features from all the segmensts for all the four tasks and
# returns y_train as (n,4), one coulum for each task. Next time extracting Xy for any particular
# task won't extract the features agains, unless you force it by setting 'redo'=True.
X_train,y_train,X_test, y_test = Subj.getXy_eeg(task=3)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
100%|##################################################|100\100|Sg - 0
Done..
100%|##################################################|43\43|Sg - 0
Done..
DataShape: (100, 84) (100,) (43, 84) (43,)
Class labels : [0 1]
# Normalization - SVM works well with normalized features
means = X_train.mean(0)
std = X_train.std(0)
X_train = (X_train-means)/std
X_test = (X_test-means)/std
# Training
clf = svm.SVC(kernel='rbf', C=1,gamma='auto')
clf.fit(X_train,y_train)
# Predition
ytp = clf.predict(X_train)
ysp = clf.predict(X_test)
# Evaluation
print('Training Accuracy:',np.mean(y_train==ytp))
print('Testing Accuracy:',np.mean(y_test==ysp))
Training Accuracy: 0.88
Testing Accuracy: 0.6046511627906976