Introduction: Getting started
In this section, we explain how to get started with the dataset and modeling. For ease all the necessary element and codes are put into one python library called - phyaat. Here we explain the functionalities that Phyaat library has with possible tuning the process of preprocessing and feature extractions. To start with a quick exmaple to preditive modeling check the Predictive Modeling tab.
For quick start with predictive modeling, check EXAMPLE CODETable of Contents
- 1. Install Library
- 2. Download dataset
- 3. Preprocessing
- 4. Extract X,y for a task Rhythmic Features
- 5. Predictive Modelling
- 6. Extracting LWR segments for extranal processing
1. Install Library
First install the python library
pip install phyaat
2. Download dataset
Once Phyaat library is installed, the dataset can be downloaded using it. You could download all the dataset together or data of one particulat subject for testing and running.
import phyaat
print('Version :' ,phyaat.__version__)
import phyaat as ph
2.1 To download dataset of subject #1
To download data set of only one subject with subject id=1 (subject=1), use following code. Here baseDir = ‘../PhyAAt_Data’ is path where data will be downloaded and stored. Make sure you have permission to write in given path.
dirPath = ph.download_data(baseDir='../PhyAAt_Data', subject=1,verbose=0,overwrite=False)
#returns a dictionary containing file names of all the subjects available in baseDir
SubID = ph.ReadFilesPath(dirPath)
# list of all the subjects in the dataset directory
print(SubID.keys())
2.2 To download dataset of all the subjects
dirPath = ph.download_data(baseDir='../PhyAAt_Data', subject=-1,verbose=0,overwrite=False)
#Check the humber of subjects are in directory - read the file path of all the subjects available
baseDir='../PhyAAt_Data' # or dirPath return path from above
#returns a dictionary containing file names of all the subjects available in baseDir
SubID = ph.ReadFilesPath(baseDir)
# list of all the subjects in the dataset directory
print(SubID.keys())
3. Preprocessing
#Creat an object holding data of a subjects
Subj = ph.Subject(SubID[1])
3.1. Filtering
Highpass filter with cut-off frrequency of 0.5Hz
This is very standard to remove any dc component and drift in the signal
#filtering with highpass filter of cutoff frequency 0.5Hz
Subj.filter_EEG(band =[0.5],btype='highpass',order=5)
Filtering with custum range of feequecy should be between 0-64Hz To analyse EEG in particulare band of frequency, such as for ERP analysis, you might need to apply for custom range of frequency band.
Lowpass filter
#filtering with lowpass filter
Subj.filter_EEG(band =[30],btype='lowpass',order=5)
Bandpass filter
#filtering with bandpass filter Theta
Subj.filter_EEG(band =[4,8],btype='bandpass',order=5)
Filter settings
#method = 'lfilter' # 'filtfilt', 'SOS'
#useRaw=False # if True, it will use raw eeg and overwirte old processed EEG
Subj.filter_EEG(band =[0.5],btype='highpass',order=5,method='lfilter',fs=128.0,verbose=0,use_joblib=False,n_jobs=-1,useRaw=False)
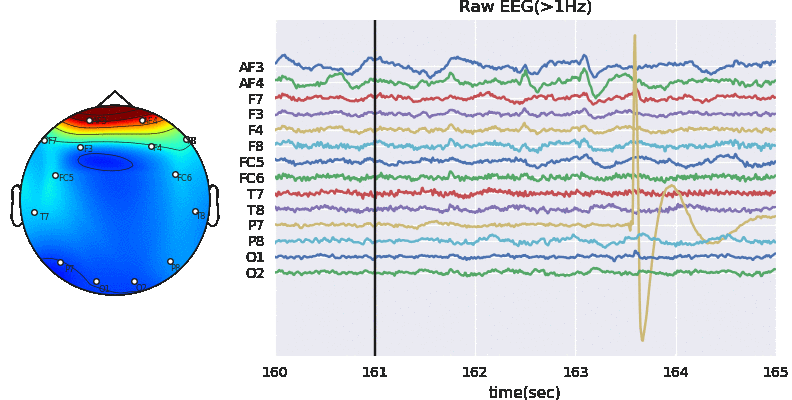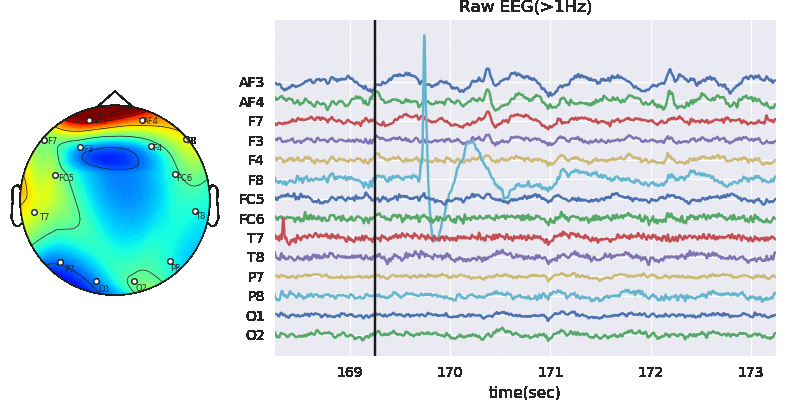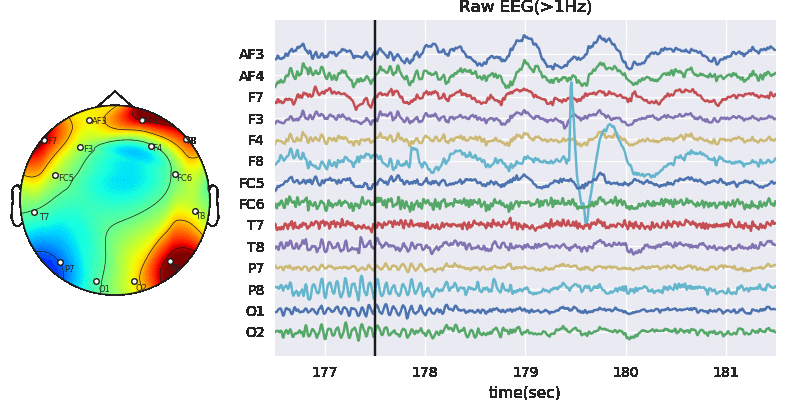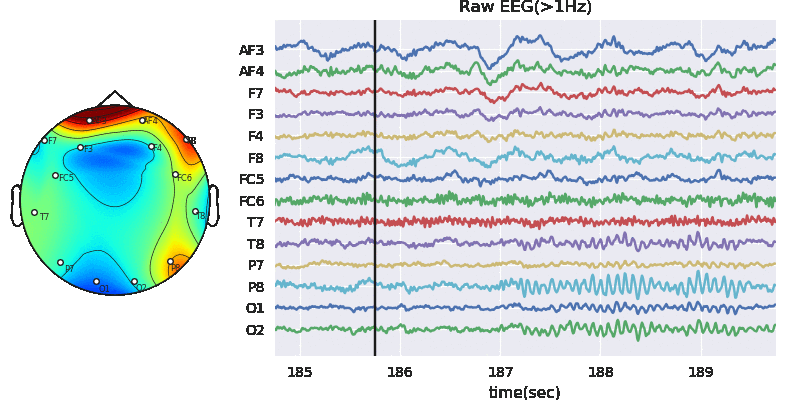
3.2 Applyting Artifact Removal Algorithm on EEG
ATAR Algorithm - Wavelet based approach (in version>0.0.2)
A wavelet based tunable algorithm
# with window size =128 (1 sec, recommonded). To save time, use winsize=128*10, 10 sec window
Subj.correct(method='ATAR',verbose=1,winsize=128, wv='db3', thr_method='ipr', OptMode='soft',beta=0.1)
# check all the parameters for ATAR
help(ph.Subject.correct)
Tune the parameters of ATAR algorithm: Mostly, we tune beta,
OptMode='elim'
beta=0.2
Subj.correct(method='ATAR',verbose=1,winsize=128, wv='db3', thr_method='ipr', OptMode=OptMode,beta=beta)
#check all the parameters here
help(ph.Subject.correct)
ICA based approach
# with window size =128 (1 sec, recommonded). To save time, use winsize=128*10, 10 sec window
Subj.correct(method='ICA',verbose=1,winsize=128)
Change parameters of ICA based artifact Removal
KurThr = 2
Corr = 0.8
ICAMed = 'extended-infomax' #picard, fastICA
Subj.correct(method='ICA',winsize=128,hopesize=None,Corr=Corr,KurThr=KurThr,
ICAMed=ICAMed,verbose=0, window=['hamming',True],
winMeth='custom')
#check all the parameters here
help(ph.Subject.correct)
4. Extract X,y for a task Rhythmic Features
4.1 Extracting Features Segment-wise
# Task 4: LWR classification
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=4)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
# Task 1: Attention Score Prediction
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=1)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nlabels :',np.unique(y_train))
# Task 2: Noise Level Predicition
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=2)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nlabels :',np.unique(y_train))
# Task 3: Semanticity Classification
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=3)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
#If features are extracted for task 1, 2 or 3 (listening segments)
# next time while extracting won't compute features again, unless redo=True
4.1 Extracting Features Window-wise
winsize=128 # 1 sec window
hopesize=32 # 0.25 shift for next window, if None, overlape is half of windowsize
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=1, features='rhythmic',
winsize=winsize, hopesize=hopesize)
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
4.2 Random split for train-test
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=1, features='rhythmic',
winsize=winsize, hopesize=hopesize,split='random')
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
4.3 Hyperparameters for feature extraction method
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=1, features='rhythmic', eSample=[0, 0],
verbose=1, redo=False, split='serial', splitAt=100, normalize=False,
log10p1=True, flat=True, filter_order=5, method='welch', window='hann',
scaling='density', detrend='constant', period_average='mean',
winsize=-1, hopesize=None)
#Check help
print(ph.Subject.getXy_eeg)
4.4 Extracting EEG Features with custom frequency bands
fBands = [[None,8],[8,24],[24,32]]
X_train,y_train, X_test,y_test = Subj.getXy_eeg(task=1, redo=True,normalize=False, log10p1=True,
flat=False, filter_order=5, filter_method='SOS', method='welch', window='hann',
scaling='density', detrend='constant', period_average='mean',
fBands=fBands, Sum=True, Mean=False, SD=False,verbose=0,
useRaw=False,redo_warn=True,use_v0=False)
#Check help
print(ph.Subject.getXy_eeg)
5. Predictive Modeling
Once you have X_train,y_train, X_test,y_test, it is easy to apply any ML or DL model to train and test. Here is a simple example of SVM. For more details on other models, check here - Predictive Modeling Examples
# Normalization - SVM works well with normalized features
means = X_train.mean(0)
std = X_train.std(0)
X_train = (X_train-means)/std
X_test = (X_test-means)/std
# Training
clf = svm.SVC(kernel='rbf', C=1,gamma='auto')
clf.fit(X_train,y_train)
# Predition
ytp = clf.predict(X_train)
ysp = clf.predict(X_test)
# Evaluation
print('Training Accuracy:',np.mean(y_train==ytp))
print('Testing Accuracy:',np.mean(y_test==ysp))
6. Extracting LWR segments for extranal processing
L,W,R, Scores, Cols = Subj.getLWR()
Check here - code for using extracting signals and processing with external libraries