Using External Libraries
In this notebook, we explain how to extract the signals preprocess with external libraries and do predictive modeling.
Table of Contents
Import Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import svm
import phyaat
print('Version :' ,phyaat.__version__)
import phyaat as ph
PhyAAt Processing lib Loaded...
Version : 0.0.2
Read the data of subject=1
dirPath = ph.download_data(baseDir='../PhyAAt_Data', subject=1,verbose=0,overwrite=False)
baseDir='../PhyAAt_Data'
SubID = ph.ReadFilesPath(baseDir)
Subj = ph.Subject(SubID[1])
Total Subjects : 1
Subj.filter_EEG(band =[0.5],btype='highpass',order=5)
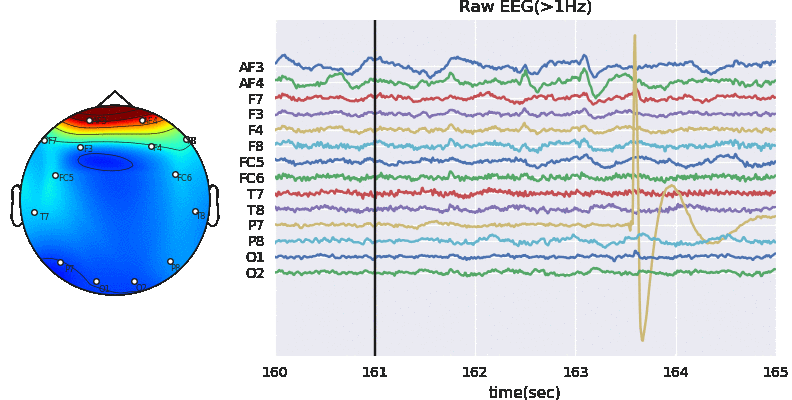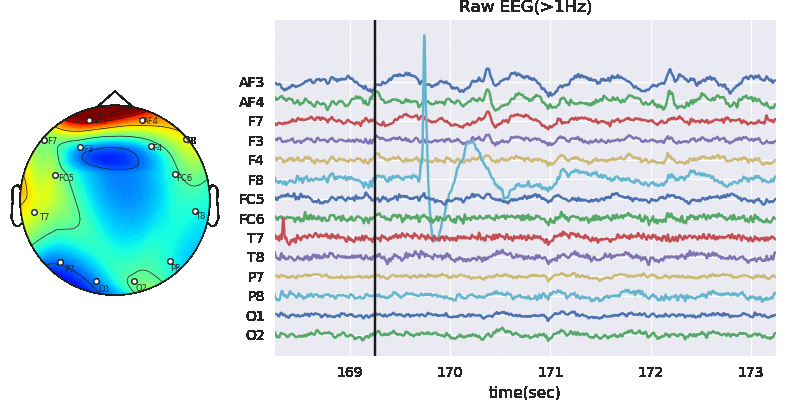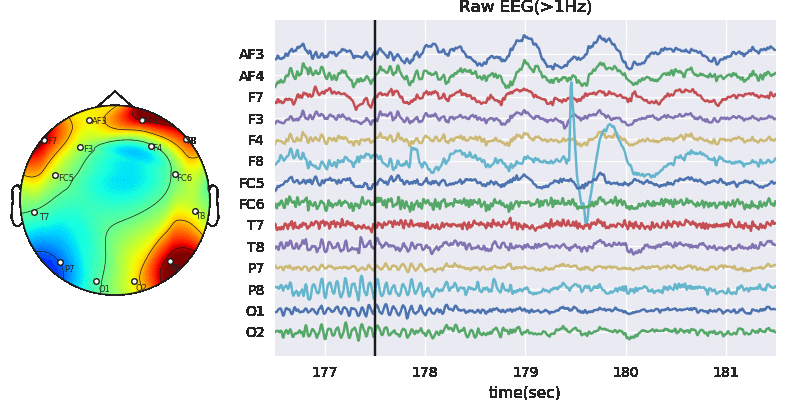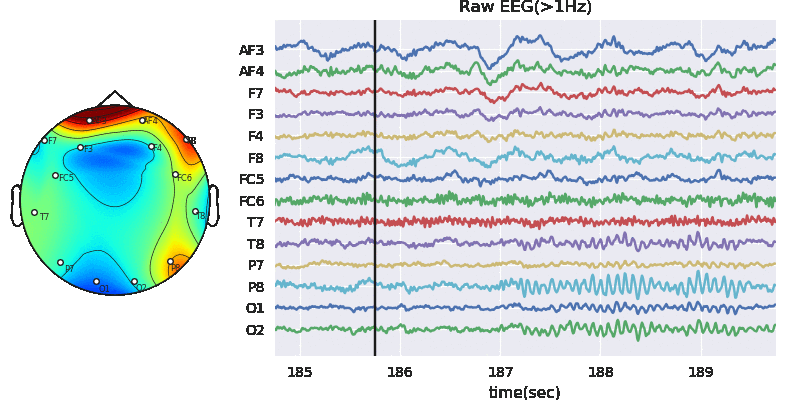
Extract Raw Signals
XE = Subj.getEEG().to_numpy()
XG = Subj.getGSR().to_numpy()
XP = Subj.getPPG().to_numpy()
plt.figure(figsize=(13,3))
plt.plot(XE[:10000,:]+np.arange(14)*100)
plt.show()
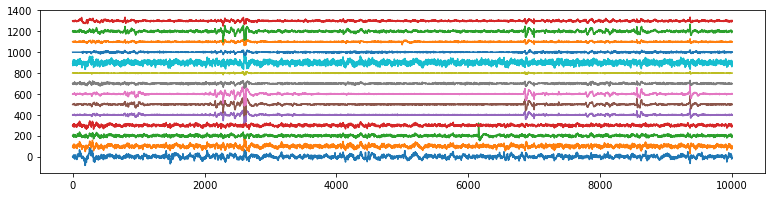
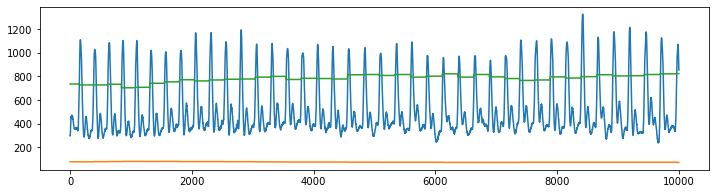
plt.figure(figsize=(12,3))
plt.plot(XG[:10000,:])
plt.show()
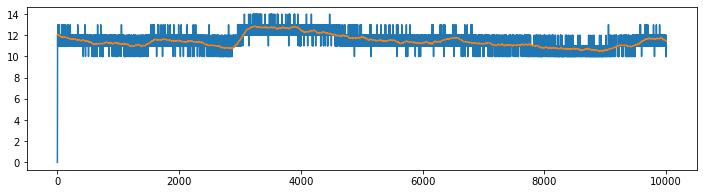
plt.figure(figsize=(12,3))
plt.plot(XP[:10000,:])
plt.show()