Feature Extraction Framework
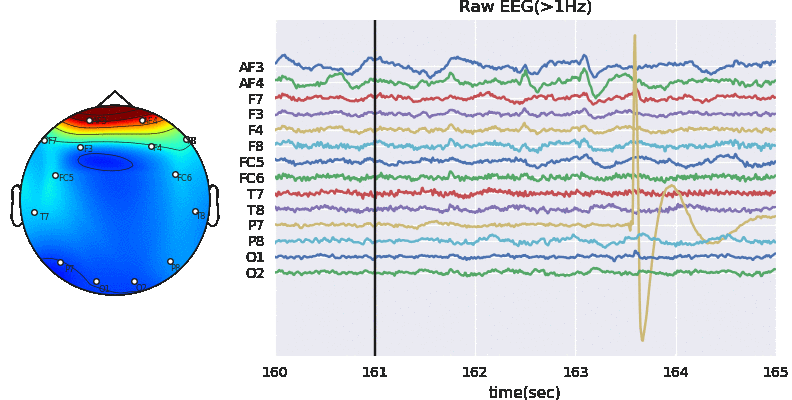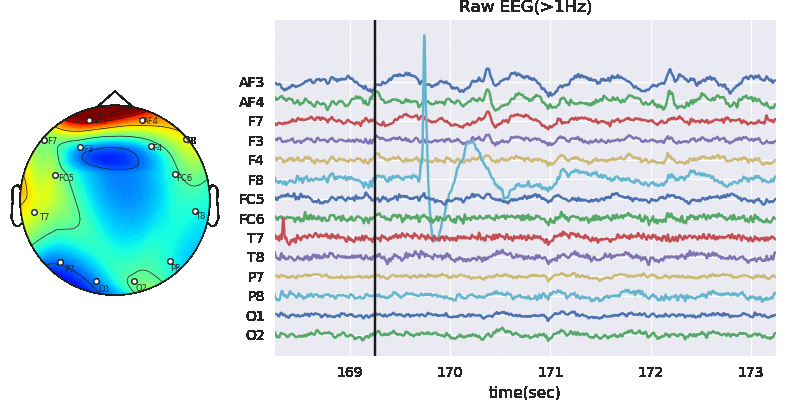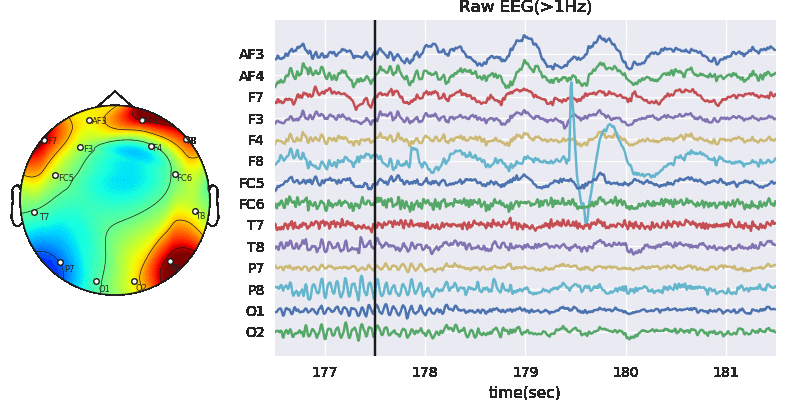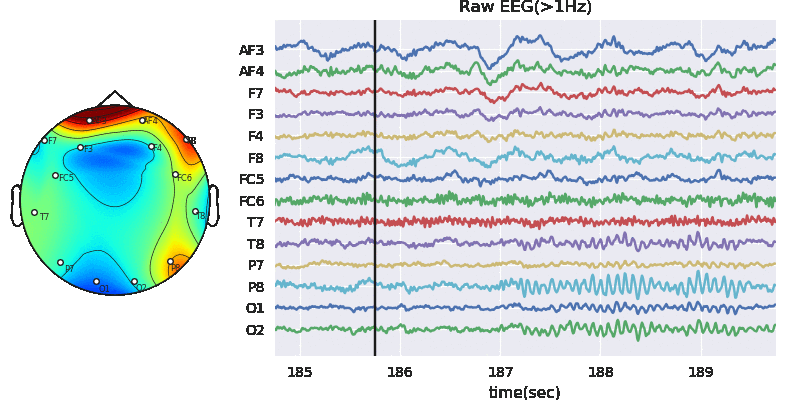
In this notebook, we explain how to extract features segment-wise and window-wise (check the paper for theoretical details). We would also explain the possible parameters to tune the feature extraction process. PhyAAt library has function to extract Rhythemic features from EEG; either segment-wise or window-wise, so we focus on only EEG feature extraction. Custom features from EEG, PPG or GSR can be done in similar fashion. Figure below shows Feature-Extraction Framework described in paper.
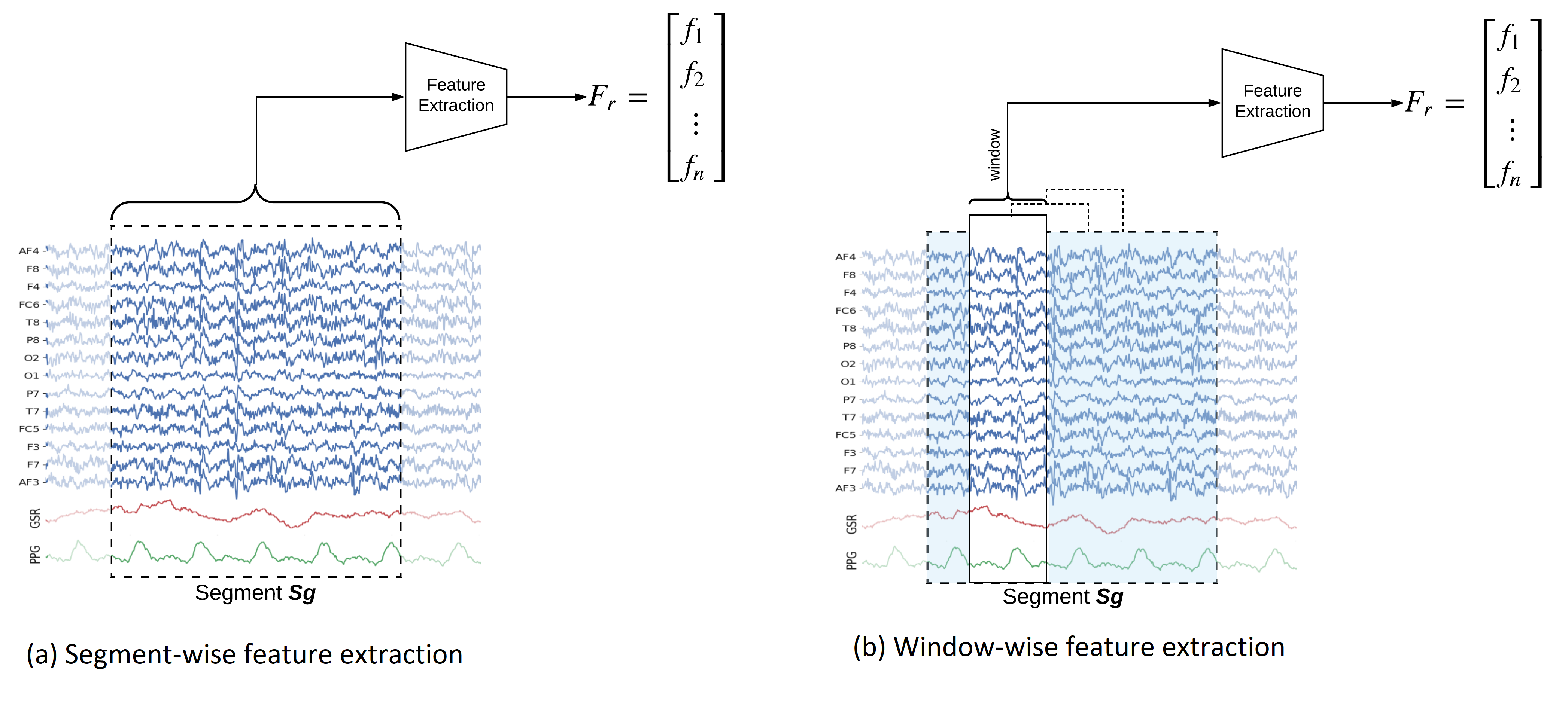
Table of Contents
Import Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import svm
import phyaat
print('Version :' ,phyaat.__version__)
import phyaat as ph
PhyAAt Processing lib Loaded...
Version : 0.0.2
Read the data of subject=1
dirPath = ph.download_data(baseDir='../PhyAAt_Data', subject=1,verbose=0,overwrite=False)
baseDir='../PhyAAt_Data'
SubID = ph.ReadFilesPath(baseDir)
Subj = ph.Subject(SubID[1])
Total Subjects : 1
Filtering
With Custum frequency range
Subj.filter_EEG(band =[0.5],btype='highpass',order=5)
Artifact removal using ICA
Tune threshold, windowsize, ICA method
KurThr = 2
Corr = 0.8
ICAMed = 'extended-infomax' #picard, fastICA
winsize=128*10 # 20sec
Subj.correct(method='ICA',winsize=winsize,hopesize=None,Corr=Corr,KurThr=KurThr,
ICAMed=ICAMed,verbose=1,
window=['hamming',True],winMeth='custom')
ICA Artifact Removal : extended-infomax
100%|###########################################################|
Feature Extraction for T4: LWR Classification
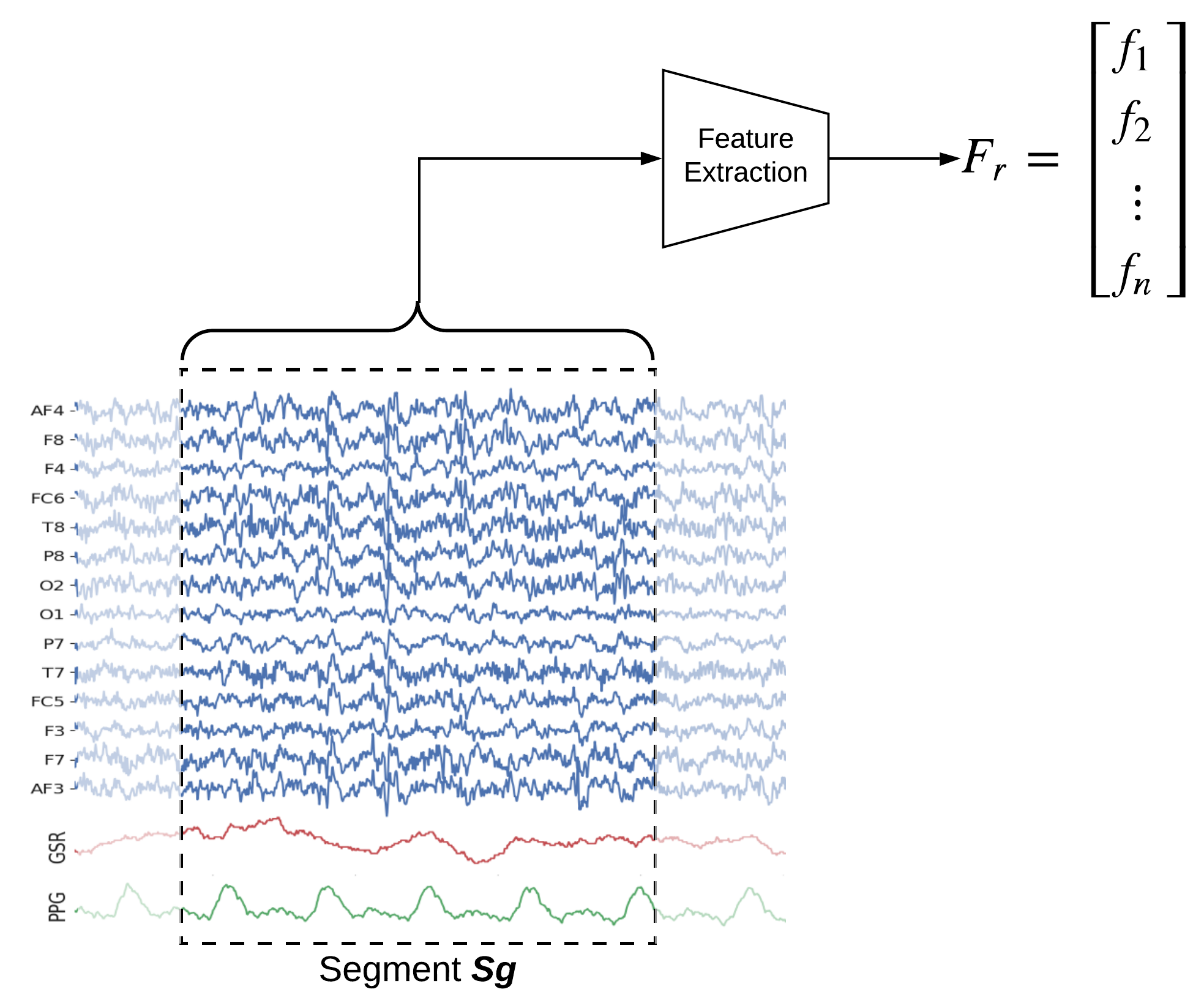
Segment-wise feature extraction
For extracting segment-wise features set winsize=-1, which will extract 84 (=6x14) features from each segment
X_train,y_train,X_test, y_test = Subj.getXy_eeg(task=4,features='rhythmic',verbose=1,
redo=False, winsize=-1,hopesize=None)
100%|##################################################|100\100|Sg - 0
Done..
100%|##################################################|100\100|Sg - 1
Done..
100%|##################################################|100\100|Sg - 2
Done..
100%|##################################################|43\43|Sg - 0
Done..
100%|##################################################|43\43|Sg - 1
Done..
100%|##################################################|43\43|Sg - 2
Done..
Shape of X_train,y_train,X_test, y_test
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
DataShape: (290, 84) (290,) (120, 84) (120,)
Class labels : [0 1 2]
# Normalization - SVM works well with normalized features
means = X_train.mean(0)
std = X_train.std(0)
X_train = (X_train-means)/std
X_test = (X_test-means)/std
# Training
clf = svm.SVC(kernel='rbf', C=1,gamma='auto')
clf.fit(X_train,y_train)
# Predition
ytp = clf.predict(X_train)
ysp = clf.predict(X_test)
# Evaluation
print('Training Accuracy:',np.mean(y_train==ytp))
print('Testing Accuracy:',np.mean(y_test==ysp))
Training Accuracy: 0.9448275862068966
Testing Accuracy: 0.85
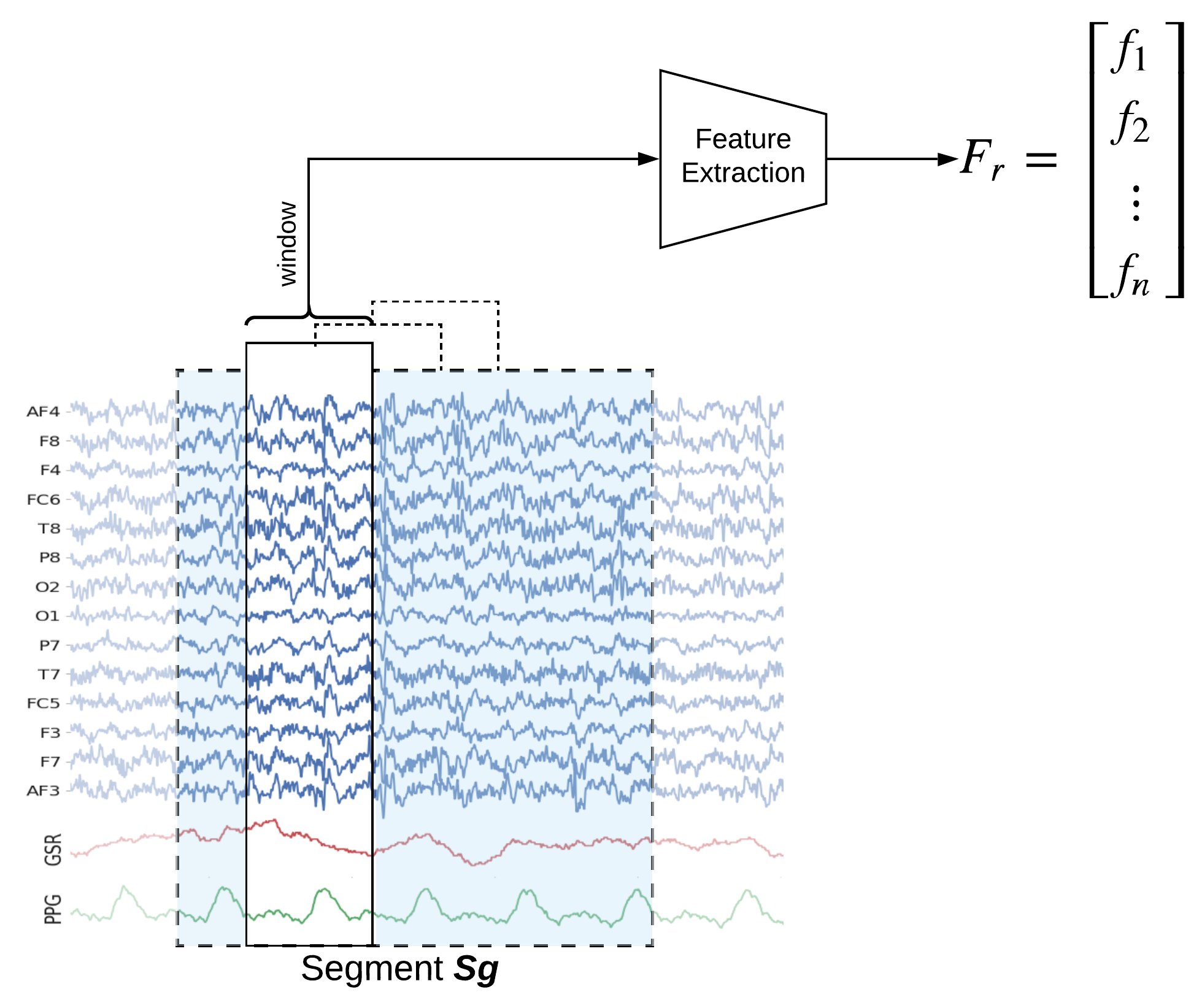
Window-wise Feature Extraction (2sec window)
For extracting Window-wise features with window size 2sec and 50% overlapping set winsize=128*2, which will extract 84 (=6x14) features from each window
X_train,y_train,X_test, y_test = Subj.getXy_eeg(task=4,features='rhythmic',
verbose=1, redo=True, winsize=128*2)
100%|##################################################|100\100|Sg - 0
Done..
100%|##################################################|100\100|Sg - 1
Done..
100%|##################################################|100\100|Sg - 2
Done..
100%|##################################################|43\43|Sg - 0
Done..
100%|##################################################|43\43|Sg - 1
Done..
100%|##################################################|43\43|Sg - 2
Done..
Notice the number of exmaples (Shape of X, y)
Shape of X_train,y_train,X_test, y_test
print('DataShape: ',X_train.shape,y_train.shape,X_test.shape, y_test.shape)
print('\nClass labels :',np.unique(y_train))
DataShape: (1382, 84) (1382,) (565, 84) (565,)
Class labels : [0 1 2]
# Normalization - SVM works well with normalized features
means = X_train.mean(0)
std = X_train.std(0)
X_train = (X_train-means)/std
X_test = (X_test-means)/std
# Training
clf = svm.SVC(kernel='rbf', C=1,gamma='auto')
clf.fit(X_train,y_train)
# Predition
ytp = clf.predict(X_train)
ysp = clf.predict(X_test)
# Evaluation
print('Training Accuracy:',np.mean(y_train==ytp))
print('Testing Accuracy:',np.mean(y_test==ysp))
Training Accuracy: 0.9261939218523878
Testing Accuracy: 0.8407079646017699
Tuning Feature Extraction process
# Subj = ph.Subject(SubID[1])
# Subj.filter_EEG(band =[0.5],btype='highpass',order=5)
# KurThr = 2
# Corr = 0.8
# ICAMed = 'extended-infomax' #picard, fastICA
# winsize=128*10 # 20sec
# Subj.correct(method='ICA',winsize=winsize,hopesize=None,Corr=Corr,KurThr=KurThr,ICAMed=ICAMed,verbose=1)
#check help to see the options
help(ph.Subject.getXy_eeg)
X_train,y_train,X_test, y_test = Subj.getXy_eeg(task=4,features='rhythmic',
verbose=1,
redo=False,
split='serial', splitAt=100,
normalize=False,log10p1=True,flat=True,
filter_order=5,method='welch',
window='hann', scaling='density',
detrend='constant',period_average='mean',
winsize=-1,hopesize=None)